Taxonomy annotations are used to indicate the taxonomy of a sequence.
A taxonomy annotation is specified as a tax=nnn field in the sequence label. Here, nnn is an integer giving the node in a taxonomy tree file. The tax=nnn field may appear anywhere in the label. It must be delimited by semi-colons, which may optionally be omitted at the end of a label, though this is not recommended. The following label has a valid taxonomy annotation:
>KR08766;tax=2034;I usually distribute FASTA files with labels in this format (the white space inside the label is a tab)::
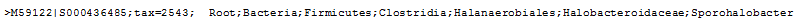
Taxonomic names are specified as
Root;Kingdom;Phylum... to the lowest classification level (usually, genus or
species). This format is compatible with the utax
command and is also accepted as input for
training the command-line version of the RDP Naive Bayesian Classifier. When
used as input to the utax command, the names are not
needed, only the tax=nnn field is required. However, it is often nice to see the
names, e.g. when reviewing hits from
usearch_global.