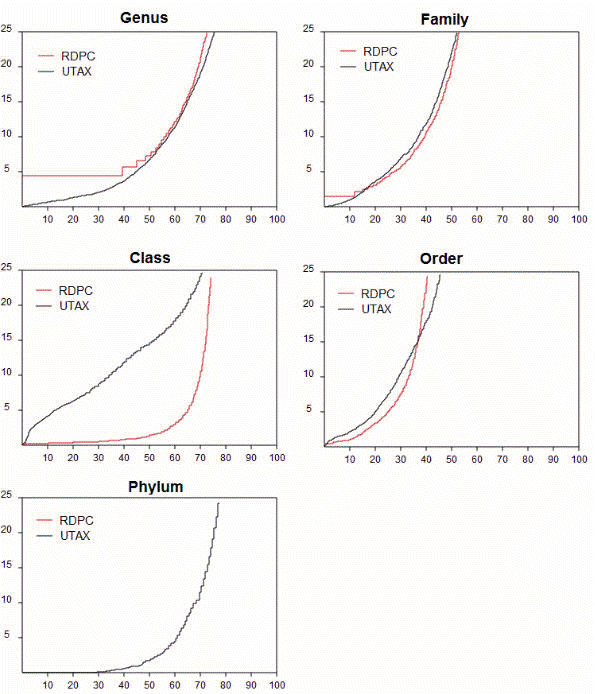
The figure below shows sensitivity (x axis) vs. error rate
(y axis) for RDP and UTAX on full-length ITS1. ITS2
is not shown; it looks similar. The method used to measure sensitivity and error
is described here:
/usearch/manual/taxonomy_validation.html
To claim that UTAX is definitively better than RDP, we would need to see that
the UTAX curve (black) is always below the RDP curve (red). This would show that
for any chosen cutoff in the RDP bootstrap, there is a P-value cutoff for UTAX
that gives higher sensitivity and lower error rate. For genus, this is true, and
for family it is mostly true at acceptable error levels (say, <5%), but the
story gets messier at higher taxonomic levels. This is the main reason I
haven't published anything yet -- this story is not a clean win, and I don't yet
understand the behavior. Why the dramatic difference between these two quite
similar algorithms at class and phylum level? UTAX is very poor compared to RDP
at class level, while UTAX does a decent job at phylum level where RDP is
useless (the first point has >25% error rate, so there is no visible RDP curve).
Note that "class level" means that the closest sequence in the reference
database is a different order or higher, it does not mean that the class level
overall is this bad -- obviously, if you get the genus or family right, all the
higher levels are also correct.
Looking at these curves, I think it is fair to say that UTAX is a better
classifier at genus and family level, and with ITS higher levels are going to be
very hard with any classifier because sequence similarity is so low.
There is another consideration that is very important -- setting cutoff values. Here, UTAX has a decisive advantage over RDP because the P-value estimates were obtained empirically from exactly these curves, so we know that 1. they accurately predict error rates, at least on this training data, and 2. the cutoffs are comparable at all levels, so e.g. if you set a 95% P-value cutoff then this will give the same error rate at each level. With the RDP classifier, the bootstrap value does not give you the error rate, and the values are not comparable at different taxonomic levels, so e.g. a 50% bootstrap cutoff might give you a 15% error rate at genus level but 30% at family level. I would prefer to have a classifier with a somewhat higher error rate if it provides informative P-values because this gives me a good estimate of the error rates for downstream analysis. With the RDP classifier, you don't get an informative estimate of error rate, so the cutoff you choose might give you higher error rates than UTAX even if lower rates are possible.